for big Data Analysis and Visualisation
Session 3 : Data Manipulation¶
Welcome to Session 3 of the Python Programming for big Data Analysis and Visualisation course. In this notebook you will find the material covered during this session.
Exercises¶
There are 14 exercises and 2 mini-projects in this session. Most will be corrected in class, and some are left for you to do.
- Exercise 1
- Exercise 2
- Exercise 3
- Exercise 4
- Exercise 5
- Exercise 6
- Exercise 7
- Exercise 8
- Exercise 9
- Exercise 10: Querying
- Exercise 11: Multiples of 5
- Exercise 12: Euclidean distance
- Exercise 13: More Titanic
- Exercise 14: Exams
- Project 1: Flower shop
- Project 2: Euro 2012
Recap: Python¶
- Scalar types:
int,float - Container types:
str,list,tuple,dict - Operators (
+,-,*,/,%) and some methods, indexing for sequences (::)str:format, find, count, replace, upper, lower, split, join;list:append, insert, remove, sort, reverse;dict:values, keys, items;
- For loops to iterate
- If clauses to make decisions based on comparison (
==,<,>) and combinations (and,or,not) - Functions to wrap up code that does something (code reuse), default and keyword arguments
- Python modules, using
importgive us access to interminable functionality
Do not forget your main resources:
- http://docs.python.org
- http://stackoverflow.com/tags/python (>1M questions)
help(...)and?...
Powering pandas (numpy)¶

Behind much of pandas' functionality lies NumPy (http://numpy.org), the main library for scientific computing in Python.
pandas: the Series data structure¶

The pandas.Series is a one-dimensional labeled array capable of holding data of any type (e.g. integer, float, object, etc.). The axis labels are collectively called index: labels need not be unique but must be immutable type (like for dict).
We will see how to:
- Create a
Series; - Query a
Series(access its elements); - Perform operations on a
Series; - Modify a
Series.
To use the pandas library, we have to import the corresponding module.
import pandas as pd
?pd.Series
Init signature: pd.Series( data=None, index=None, dtype=None, name=None, copy=False, fastpath=False, ) Docstring: One-dimensional ndarray with axis labels (including time series). Labels need not be unique but must be a hashable type. The object supports both integer- and label-based indexing and provides a host of methods for performing operations involving the index. Statistical methods from ndarray have been overridden to automatically exclude missing data (currently represented as NaN). Operations between Series (+, -, /, *, **) align values based on their associated index values-- they need not be the same length. The result index will be the sorted union of the two indexes. Parameters ---------- data : array-like, Iterable, dict, or scalar value Contains data stored in Series. .. versionchanged:: 0.23.0 If data is a dict, argument order is maintained for Python 3.6 and later. index : array-like or Index (1d) Values must be hashable and have the same length as `data`. Non-unique index values are allowed. Will default to RangeIndex (0, 1, 2, ..., n) if not provided. If both a dict and index sequence are used, the index will override the keys found in the dict. dtype : str, numpy.dtype, or ExtensionDtype, optional Data type for the output Series. If not specified, this will be inferred from `data`. See the :ref:`user guide <basics.dtypes>` for more usages. name : str, optional The name to give to the Series. copy : bool, default False Copy input data. File: ~/opt/anaconda3/lib/python3.8/site-packages/pandas/core/series.py Type: type Subclasses: SubclassedSeries
Creating a Series¶
Either using a dict: then keys will become the index.
menu_dict = {
"salad": 8.0,
"water": 1.5,
"muffin": 4.5,
"coffee": 2.0
}
menu = pd.Series(menu_dict)
menu
salad 8.0 water 1.5 muffin 4.5 coffee 2.0 dtype: float64
Or using a list: then default index will be assigned (integers starting at 0). An index can be specified using the index keyword argument.
animals = pd.Series(["Tiger", "Bear", "Moose"]) # default index = [0, 1, 2]
animals = pd.Series(["Tiger", "Bear", "Moose"], index=[1, 2, 3])
animals
1 Tiger 2 Bear 3 Moose dtype: object
The index of the Series can be accessed with Series.index.
menu.index
Index(['salad', 'water', 'muffin', 'coffee'], dtype='object')
animals.index
RangeIndex(start=0, stop=3, step=1)
x_dict = {
"a": 0,
"b": 1,
"c": 2,
"d": 3,
"e": 4,
"f": 5
}
x = pd.Series(x_dict)
x
a 0 b 1 c 2 d 3 e 4 f 5 dtype: int64
x = pd.Series(range(6), index=list("abcdef"))
x
a 0 b 1 c 2 d 3 e 4 f 5 dtype: int64
Querying a Series¶
A Series is ordered (like a list) and has labels (i.e. the index, like keys for a dict).
We query a Series using .loc[] and .iloc[].
.ilocusing positions (similar to alist);.locusing labels (similar to adict).
Note that all indexing methods we saw for str and list work here (e.g. negative indices, ranges and slices). We can also use a list to get multiple specified elements in a precise order.
menu
salad 8.0 water 1.5 muffin 4.5 coffee 2.0 dtype: float64
menu.loc["salad"]
8.0
menu.iloc[0]
8.0
menu.iloc[::2]
salad 8.0 muffin 4.5 dtype: float64
menu.loc[ ["salad", "water"] ] # elements are returned in the order of the list
salad 8.0 water 1.5 dtype: float64
Exercise 2¶
Extract from the x Series created above (Exercise 1) the elements as specified below:
- the first element
- the last element
- the element with label "d"
- every second element starting at the beginning
- third and fifth elements
- fourth, second and third elements, in this order
x.iloc[0]
x.iloc[-1]
x.loc["d"]
x.iloc[::2]
x.iloc[ [2, 4] ]
x.loc[ ["c", "e"] ] # alternative solution to 4.
x.iloc[ [3, 1, 2] ]
d 3 b 1 c 2 dtype: int64
Series operations¶
Although iterating through a Series is possible, vectorised operations should be prefered (when applicable), for both readability and performance.
Operators that work on scalars (e.g. +,-,*,/,**,%,etc...) also work on Series by applying the operation to each value.
Series also have a number of methods that perform operations (e.g. sum, min, max, mean, median, mode, std, var, sem, abs, round).
# PLEASE DO NOT DO THIS
for element in menu:
print(element)
8.0 1.5 4.5 2.0
# PLEASE DO NOT DO THIS
for label, value in menu.items():
print(f"{label} costs {value} dollars")
salad costs 8.0 dollars water costs 1.5 dollars muffin costs 4.5 dollars coffee costs 2.0 dollars
For example, let's say we want to see the menu with prices augmented by 1.
# Using a for loop
for element in menu:
print(element + 1)
9.0 2.5 5.5 3.0
# Using a vectorised operation: FASTER and MORE READABLE
menu_expensive = menu + 1
menu_expensive
salad 9.0 water 2.5 muffin 5.5 coffee 3.0 dtype: float64
Another example: sum all prices in the menu.
# Using a for loop
total = 0
for price in menu:
total = total + price
total
16.0
# Using a vectorised operation: FASTER and MORE READABLE
menu.sum()
16.0
Exercise 3¶
Calculate the following values starting from the x Series:
- $x + 2$
- $x^2$
- $3x^3 + 2x^2 - 5x + 7$
- The sum of $x^3$
x + 2
x ** 2
3 * x**3 + 2 * x**2 - 5 * x + 7
x_cubes = x**3
x_cubes.sum()
(x**3).sum() # more concisely
225
Modifying Series¶
We can assign to values in a Series just like for a list or dict. NOTE that all indexing methods work.
We can add another Series to a Series using the pd.concat method.
menu.loc["salad"] = 7.0 # as in dict
menu.iloc[0] = 6.5 # as in list
menu.iloc[::2] = 1.0
menu.iloc[::2] = [7.0, 4.5]
menu.loc["pizza"] = 9.5
menu
salad 7.0 water 1.5 muffin 4.5 coffee 2.0 pizza 9.5 dtype: float64
new_menu_items = pd.Series({"chips": 3.5})
new_menu = pd.concat((menu, new_menu_items))
new_menu
salad 7.0 water 1.5 muffin 4.5 coffee 2.0 pizza 9.5 chips 3.5 dtype: float64
Exercise 4¶
Modify the x Series in the following ways :
- Add the next logical element;
- Change the value of
cto 3; - Modify values
d,eandfto 4; - Modify values
d,eandfto 6, 5, 4, respectively.
# x = pd.concat((x, pd.Series({"g": 6})))
x.loc["g"] = 6
x.loc["c"] = 3
x.loc[ ["d", "e", "f"] ] = 4
x.loc["d":"f"] = 4 # alternative solution
x.loc["d":"f"] = [6, 5, 4]
x
a 0 b 1 c 3 d 6 e 5 f 4 g 6 dtype: int64
pandas: the DataFrame data structure¶
The pandas.DataFrame is a two-dimensional data structure, containing data aligned in a tabular fashion in rows and columns.
| name | sex | age | fare | survived |
|---|---|---|---|---|
| Allen | male | 35 | 8.05 | 0 |
| Braund | male | 22 | 7.25 | 0 |
| Cumings | female | 38 | 71.28 | 1 |
| Futrelle | female | 35 | 53.10 | 1 |
| Futrelle | male | 37 | 53.10 | 0 |
| Heikkinen | female | 26 | 7.90 | 1 |
We will see how to:
- Create a
DataFrame; - Query a
DataFrame(access its elements); - Modify a
DataFrame; - Read and write
DataFrames from/to files.
?pd.DataFrame
Init signature: pd.DataFrame( data=None, index: 'Axes | None' = None, columns: 'Axes | None' = None, dtype: 'Dtype | None' = None, copy: 'bool | None' = None, ) Docstring: Two-dimensional, size-mutable, potentially heterogeneous tabular data. Data structure also contains labeled axes (rows and columns). Arithmetic operations align on both row and column labels. Can be thought of as a dict-like container for Series objects. The primary pandas data structure. Parameters ---------- data : ndarray (structured or homogeneous), Iterable, dict, or DataFrame Dict can contain Series, arrays, constants, dataclass or list-like objects. If data is a dict, column order follows insertion-order. If a dict contains Series which have an index defined, it is aligned by its index. .. versionchanged:: 0.25.0 If data is a list of dicts, column order follows insertion-order. index : Index or array-like Index to use for resulting frame. Will default to RangeIndex if no indexing information part of input data and no index provided. columns : Index or array-like Column labels to use for resulting frame when data does not have them, defaulting to RangeIndex(0, 1, 2, ..., n). If data contains column labels, will perform column selection instead. dtype : dtype, default None Data type to force. Only a single dtype is allowed. If None, infer. copy : bool or None, default None Copy data from inputs. For dict data, the default of None behaves like ``copy=True``. For DataFrame or 2d ndarray input, the default of None behaves like ``copy=False``. .. versionchanged:: 1.3.0 See Also -------- DataFrame.from_records : Constructor from tuples, also record arrays. DataFrame.from_dict : From dicts of Series, arrays, or dicts. read_csv : Read a comma-separated values (csv) file into DataFrame. read_table : Read general delimited file into DataFrame. read_clipboard : Read text from clipboard into DataFrame. Examples -------- Constructing DataFrame from a dictionary. >>> d = {'col1': [1, 2], 'col2': [3, 4]} >>> df = pd.DataFrame(data=d) >>> df col1 col2 0 1 3 1 2 4 Notice that the inferred dtype is int64. >>> df.dtypes col1 int64 col2 int64 dtype: object To enforce a single dtype: >>> df = pd.DataFrame(data=d, dtype=np.int8) >>> df.dtypes col1 int8 col2 int8 dtype: object Constructing DataFrame from a dictionary including Series: >>> d = {'col1': [0, 1, 2, 3], 'col2': pd.Series([2, 3], index=[2, 3])} >>> pd.DataFrame(data=d, index=[0, 1, 2, 3]) col1 col2 0 0 NaN 1 1 NaN 2 2 2.0 3 3 3.0 Constructing DataFrame from numpy ndarray: >>> df2 = pd.DataFrame(np.array([[1, 2, 3], [4, 5, 6], [7, 8, 9]]), ... columns=['a', 'b', 'c']) >>> df2 a b c 0 1 2 3 1 4 5 6 2 7 8 9 Constructing DataFrame from a numpy ndarray that has labeled columns: >>> data = np.array([(1, 2, 3), (4, 5, 6), (7, 8, 9)], ... dtype=[("a", "i4"), ("b", "i4"), ("c", "i4")]) >>> df3 = pd.DataFrame(data, columns=['c', 'a']) ... >>> df3 c a 0 3 1 1 6 4 2 9 7 Constructing DataFrame from dataclass: >>> from dataclasses import make_dataclass >>> Point = make_dataclass("Point", [("x", int), ("y", int)]) >>> pd.DataFrame([Point(0, 0), Point(0, 3), Point(2, 3)]) x y 0 0 0 1 0 3 2 2 3 File: ~/opt/anaconda3/lib/python3.9/site-packages/pandas/core/frame.py Type: type Subclasses: SubclassedDataFrame
Creating a DataFrame¶
There are several ways to create a DataFrame. A simple one is to create a dict of columns: each value is a list of the same size. More often, we will read DataFrames from a file (see below).
DataFrame structure¶
By default, pandas creates an index of integers. We can assign a column to be the index using the set_index method. NOTE that set_index returns a modifed DataFrame. We can set back the default index by using reset_index; as set_index, it does not modify the DataFrame.
tit_dict = {
"name": ["Allen", "Braund", "Cumings", "Futrelle", "Futrelle", "Heikkinen"],
"sex": ["male", "male", "female", "female", "male", "female"],
"age": [35, 22, 38, 35, 37, 26],
"fare": [8.05, 7.25, 71.28, 53.1, 53.1, 7.9],
"survived": [0, 0, 1, 1, 0, 1],
}
titanic = pd.DataFrame(tit_dict)
titanic.set_index("name") # returns the modified DataFrame ! has NOT modified titanic
| sex | age | fare | survived | |
|---|---|---|---|---|
| name | ||||
| Allen | male | 35 | 8.05 | 0 |
| Braund | male | 22 | 7.25 | 0 |
| Cumings | female | 38 | 71.28 | 1 |
| Futrelle | female | 35 | 53.10 | 1 |
| Futrelle | male | 37 | 53.10 | 0 |
| Heikkinen | female | 26 | 7.90 | 1 |
titanic.index
RangeIndex(start=0, stop=6, step=1)
titanic = titanic.set_index("name")
titanic.index
Index(['Allen', 'Braund', 'Cumings', 'Futrelle', 'Futrelle', 'Heikkinen'], dtype='object', name='name')
titanic.columns
Index(['sex', 'age', 'fare', 'survived'], dtype='object')
titanic.reset_index() # returns the modified DataFrame ! has NOT modified titanic
| name | sex | age | fare | survived | |
|---|---|---|---|---|---|
| 0 | Allen | male | 35 | 8.05 | 0 |
| 1 | Braund | male | 22 | 7.25 | 0 |
| 2 | Cumings | female | 38 | 71.28 | 1 |
| 3 | Futrelle | female | 35 | 53.10 | 1 |
| 4 | Futrelle | male | 37 | 53.10 | 0 |
| 5 | Heikkinen | female | 26 | 7.90 | 1 |
Exploring a DataFrame¶
We can explore a DataFrame using:
head(): lists the first 10 rows of theDataFrame;info(): gives an overview of theDataFrame, in particular sized anddtypes;describe(): gives a "statistical" description of quantitative columns (intandfloat).
titanic.head()
| sex | age | fare | survived | |
|---|---|---|---|---|
| name | ||||
| Allen | male | 35 | 8.05 | 0 |
| Braund | male | 22 | 7.25 | 0 |
| Cumings | female | 38 | 71.28 | 1 |
| Futrelle | female | 35 | 53.10 | 1 |
| Futrelle | male | 37 | 53.10 | 0 |
titanic.info()
<class 'pandas.core.frame.DataFrame'> Index: 6 entries, Allen to Heikkinen Data columns (total 4 columns): # Column Non-Null Count Dtype --- ------ -------------- ----- 0 sex 6 non-null object 1 age 6 non-null int64 2 fare 6 non-null float64 3 survived 6 non-null int64 dtypes: float64(1), int64(2), object(1) memory usage: 412.0+ bytes
titanic.describe()
| age | fare | survived | |
|---|---|---|---|
| count | 6.000000 | 6.000000 | 6.000000 |
| mean | 32.166667 | 33.446667 | 0.500000 |
| std | 6.554896 | 28.940478 | 0.547723 |
| min | 22.000000 | 7.250000 | 0.000000 |
| 25% | 28.250000 | 7.937500 | 0.000000 |
| 50% | 35.000000 | 30.575000 | 0.500000 |
| 75% | 36.500000 | 53.100000 | 1.000000 |
| max | 38.000000 | 71.280000 | 1.000000 |
Querying a DataFrame¶
Similarly to Series, we can retrieve values from a DataFrame using .loc[] (for labels) and .iloc[] (for positions).
According to what we query, pandas will return:
- scalar if there is only a single value to return;
Seriesif there is a 1D set to return;DataFrameif there is a 2D set to return;
For DataFrames, .loc[] and .iloc[] optionally accept two indices separated by comma. The first index is for rows, the second for columns. We can use the : character to indicate we want all rows or columns.
Remember: [ ROWS, COLUMNS ].
titanic
| sex | age | fare | survived | |
|---|---|---|---|---|
| name | ||||
| Allen | male | 35 | 8.05 | 0 |
| Braund | male | 22 | 7.25 | 0 |
| Cumings | female | 38 | 71.28 | 1 |
| Futrelle | female | 35 | 53.10 | 1 |
| Futrelle | male | 37 | 53.10 | 0 |
| Heikkinen | female | 26 | 7.90 | 1 |
titanic.iloc[0]
sex male age 35 fare 8.05 survived 0 Name: Allen, dtype: object
titanic.loc["Allen"]
sex male age 35 fare 8.05 survived 0 Name: Allen, dtype: object
titanic.loc["Futrelle"]
| sex | age | fare | survived | |
|---|---|---|---|---|
| name | ||||
| Futrelle | female | 35 | 53.1 | 1 |
| Futrelle | male | 37 | 53.1 | 0 |
titanic.iloc[0, 0]
'male'
titanic.iloc[0:4, 0] # All indexing tricks that work for list and str also work for pd.Series and pd.DataFrame !
name Allen male Braund male Cumings female Futrelle female Name: sex, dtype: object
titanic.loc[:, "sex"] # REMEMBER: [ROWS, COLUMNS]
name Allen male Braund male Cumings female Futrelle female Futrelle male Heikkinen female Name: sex, dtype: object
titanic.loc["Allen", "sex"] # returns a scalar (just a string)
'male'
titanic.loc["Allen", ["sex", "age"]] # returns a Series (1D set of data)
sex male age 35 Name: Allen, dtype: object
titanic.loc["Futrelle", ["sex", "age"]] # returns a DataFrame (2D set of data)
| sex | age | |
|---|---|---|
| name | ||
| Futrelle | female | 35 |
| Futrelle | male | 37 |
Exercise 5¶
Extract from the titanic DataFrame the elements as specified below:
- The last column;
- The
farepayed by Ms Cumings; - The
ageof Mr Braund; - The last row;
- Just
sexandageof all passengers; - The
ageof the last passenger; - Passengers who have
ageof 35 (HINT:set_index()); - The
farepayed by the passengers that survived (HINT:set_index()).
# REMEMBER:
# .loc[ROWS, COLUMNS]
# .iloc[ROWS, COLUMNS]
titanic.iloc[:, -1]
titanic.loc["Cumings", "fare"]
titanic.loc["Braund", "age"]
titanic.iloc[-1, :]
titanic.iloc[-1]
titanic.loc[:, ["sex", "age"]]
titanic.iloc[-1].loc["age"]
titanic.set_index("age").loc[35]
titanic.set_index("survived").loc[1, "fare"]
survived 1 71.28 1 53.10 1 7.90 Name: fare, dtype: float64
Exercise 6¶
Use the titanic DataFrame to calculate the following values:
- The total
farepayed by all these passengers (HINT:Series.sum()); - The total
farepayed by Mr and Ms Futrelle; - The number of passengers that survived;
- The mean
ageof all passengers; - The mean
farepayed by male passengers (HINT:set_index());
titanic.loc[:, "fare"].sum()
titanic.loc["Futrelle", "fare"].sum()
titanic.loc[:, "survived"].sum()
titanic.loc[:, "age"].mean()
titanic.set_index("sex").loc["male", "fare"].mean()
22.8
Modifying a DataFrame¶
Similarly to Series, we can assign to specific values in a DataFrame. NOTE that all indexing methods work.
titanic.loc["Allen", "age"] = 36
# etc...
The following commands would work, but I'm not executing them to preserve my Titanic Disaster dataset.
Can you predict what they will do ?
titanic.loc[:, "age"] = 1
titanic.loc[:, "age"] = [1,2,3,4,5,6]
A new column can be added by using .assign(), or .loc[] (just like Series).
titanic.loc[:, "visa"] = False # add a column called "visa"
titanic.assign(visa=True)
| sex | age | fare | survived | visa | |
|---|---|---|---|---|---|
| name | |||||
| Allen | male | 35 | 8.05 | 0 | True |
| Braund | male | 22 | 7.25 | 0 | True |
| Cumings | female | 38 | 71.28 | 1 | True |
| Futrelle | female | 35 | 53.10 | 1 | True |
| Futrelle | male | 37 | 53.10 | 0 | True |
| Heikkinen | female | 26 | 7.90 | 1 | True |
titanic.loc["Cumings", "visa"] = True
Exercise 7¶
Modify the titanic DataFrame as specified below:
- Set the
ageof Ms Heikkinen to 28; - Mr Braund is found alive (he was hiding in Panama): change his record accordingly;
- Add a new column
portwith value "Southampton" for all passengers; - Change the value of
portto "Queensland" for the Futrelle couple.
titanic
| sex | age | fare | survived | |
|---|---|---|---|---|
| name | ||||
| Allen | male | 35 | 8.05 | 0 |
| Braund | male | 22 | 7.25 | 0 |
| Cumings | female | 38 | 71.28 | 1 |
| Futrelle | female | 35 | 53.10 | 1 |
| Futrelle | male | 37 | 53.10 | 0 |
| Heikkinen | female | 26 | 7.90 | 1 |
titanic.loc["Heikkinen", "age"] = 28
titanic.loc["Braund", "survived"] = 1
titanic.loc[:, "port"] = "Southampton"
titanic.loc["Futrelle", "port"] = "Queensland"
Reading and writing Dataframes¶
Data is often transported in files. One of the strenghts of pandas is its ability to read and write data files in a large number of formats.
The CSV Format¶
One of the most used formats to store data in a file is the CSV format (Comma-Separated Values).
CSV files are very simple: Each line is a row, with values separated by commas:
column1,column2,column3
value1,value2,value3
By convention, the first row is often a header containing column names, and the first column is the index of the row.
You can find an example CSV file here:
https://marcopasi.github.io/physenbio_pyDAV/data/titanic_tiny.csv
pandas reads CSV files using the pd.read_csv() method, which returns a DataFrame. DataFrames can be written to CSV using the to_csv() method.
titanic = pd.read_csv("https://marcopasi.github.io/physenbio_pyDAV/data/titanic_tiny.csv")
titanic = titanic.set_index("name")
?pd.read_csv
Signature: pd.read_csv( filepath_or_buffer: Union[str, pathlib.Path, IO[~AnyStr]], sep=',', delimiter=None, header='infer', names=None, index_col=None, usecols=None, squeeze=False, prefix=None, mangle_dupe_cols=True, dtype=None, engine=None, converters=None, true_values=None, false_values=None, skipinitialspace=False, skiprows=None, skipfooter=0, nrows=None, na_values=None, keep_default_na=True, na_filter=True, verbose=False, skip_blank_lines=True, parse_dates=False, infer_datetime_format=False, keep_date_col=False, date_parser=None, dayfirst=False, cache_dates=True, iterator=False, chunksize=None, compression='infer', thousands=None, decimal: str = '.', lineterminator=None, quotechar='"', quoting=0, doublequote=True, escapechar=None, comment=None, encoding=None, dialect=None, error_bad_lines=True, warn_bad_lines=True, delim_whitespace=False, low_memory=True, memory_map=False, float_precision=None, ) Docstring: Read a comma-separated values (csv) file into DataFrame. Also supports optionally iterating or breaking of the file into chunks. Additional help can be found in the online docs for `IO Tools <https://pandas.pydata.org/pandas-docs/stable/user_guide/io.html>`_. Parameters ---------- filepath_or_buffer : str, path object or file-like object Any valid string path is acceptable. The string could be a URL. Valid URL schemes include http, ftp, s3, gs, and file. For file URLs, a host is expected. A local file could be: file://localhost/path/to/table.csv. If you want to pass in a path object, pandas accepts any ``os.PathLike``. By file-like object, we refer to objects with a ``read()`` method, such as a file handler (e.g. via builtin ``open`` function) or ``StringIO``. sep : str, default ',' Delimiter to use. If sep is None, the C engine cannot automatically detect the separator, but the Python parsing engine can, meaning the latter will be used and automatically detect the separator by Python's builtin sniffer tool, ``csv.Sniffer``. In addition, separators longer than 1 character and different from ``'\s+'`` will be interpreted as regular expressions and will also force the use of the Python parsing engine. Note that regex delimiters are prone to ignoring quoted data. Regex example: ``'\r\t'``. delimiter : str, default ``None`` Alias for sep. header : int, list of int, default 'infer' Row number(s) to use as the column names, and the start of the data. Default behavior is to infer the column names: if no names are passed the behavior is identical to ``header=0`` and column names are inferred from the first line of the file, if column names are passed explicitly then the behavior is identical to ``header=None``. Explicitly pass ``header=0`` to be able to replace existing names. The header can be a list of integers that specify row locations for a multi-index on the columns e.g. [0,1,3]. Intervening rows that are not specified will be skipped (e.g. 2 in this example is skipped). Note that this parameter ignores commented lines and empty lines if ``skip_blank_lines=True``, so ``header=0`` denotes the first line of data rather than the first line of the file. names : array-like, optional List of column names to use. If the file contains a header row, then you should explicitly pass ``header=0`` to override the column names. Duplicates in this list are not allowed. index_col : int, str, sequence of int / str, or False, default ``None`` Column(s) to use as the row labels of the ``DataFrame``, either given as string name or column index. If a sequence of int / str is given, a MultiIndex is used. Note: ``index_col=False`` can be used to force pandas to *not* use the first column as the index, e.g. when you have a malformed file with delimiters at the end of each line. usecols : list-like or callable, optional Return a subset of the columns. If list-like, all elements must either be positional (i.e. integer indices into the document columns) or strings that correspond to column names provided either by the user in `names` or inferred from the document header row(s). For example, a valid list-like `usecols` parameter would be ``[0, 1, 2]`` or ``['foo', 'bar', 'baz']``. Element order is ignored, so ``usecols=[0, 1]`` is the same as ``[1, 0]``. To instantiate a DataFrame from ``data`` with element order preserved use ``pd.read_csv(data, usecols=['foo', 'bar'])[['foo', 'bar']]`` for columns in ``['foo', 'bar']`` order or ``pd.read_csv(data, usecols=['foo', 'bar'])[['bar', 'foo']]`` for ``['bar', 'foo']`` order. If callable, the callable function will be evaluated against the column names, returning names where the callable function evaluates to True. An example of a valid callable argument would be ``lambda x: x.upper() in ['AAA', 'BBB', 'DDD']``. Using this parameter results in much faster parsing time and lower memory usage. squeeze : bool, default False If the parsed data only contains one column then return a Series. prefix : str, optional Prefix to add to column numbers when no header, e.g. 'X' for X0, X1, ... mangle_dupe_cols : bool, default True Duplicate columns will be specified as 'X', 'X.1', ...'X.N', rather than 'X'...'X'. Passing in False will cause data to be overwritten if there are duplicate names in the columns. dtype : Type name or dict of column -> type, optional Data type for data or columns. E.g. {'a': np.float64, 'b': np.int32, 'c': 'Int64'} Use `str` or `object` together with suitable `na_values` settings to preserve and not interpret dtype. If converters are specified, they will be applied INSTEAD of dtype conversion. engine : {'c', 'python'}, optional Parser engine to use. The C engine is faster while the python engine is currently more feature-complete. converters : dict, optional Dict of functions for converting values in certain columns. Keys can either be integers or column labels. true_values : list, optional Values to consider as True. false_values : list, optional Values to consider as False. skipinitialspace : bool, default False Skip spaces after delimiter. skiprows : list-like, int or callable, optional Line numbers to skip (0-indexed) or number of lines to skip (int) at the start of the file. If callable, the callable function will be evaluated against the row indices, returning True if the row should be skipped and False otherwise. An example of a valid callable argument would be ``lambda x: x in [0, 2]``. skipfooter : int, default 0 Number of lines at bottom of file to skip (Unsupported with engine='c'). nrows : int, optional Number of rows of file to read. Useful for reading pieces of large files. na_values : scalar, str, list-like, or dict, optional Additional strings to recognize as NA/NaN. If dict passed, specific per-column NA values. By default the following values are interpreted as NaN: '', '#N/A', '#N/A N/A', '#NA', '-1.#IND', '-1.#QNAN', '-NaN', '-nan', '1.#IND', '1.#QNAN', '<NA>', 'N/A', 'NA', 'NULL', 'NaN', 'n/a', 'nan', 'null'. keep_default_na : bool, default True Whether or not to include the default NaN values when parsing the data. Depending on whether `na_values` is passed in, the behavior is as follows: * If `keep_default_na` is True, and `na_values` are specified, `na_values` is appended to the default NaN values used for parsing. * If `keep_default_na` is True, and `na_values` are not specified, only the default NaN values are used for parsing. * If `keep_default_na` is False, and `na_values` are specified, only the NaN values specified `na_values` are used for parsing. * If `keep_default_na` is False, and `na_values` are not specified, no strings will be parsed as NaN. Note that if `na_filter` is passed in as False, the `keep_default_na` and `na_values` parameters will be ignored. na_filter : bool, default True Detect missing value markers (empty strings and the value of na_values). In data without any NAs, passing na_filter=False can improve the performance of reading a large file. verbose : bool, default False Indicate number of NA values placed in non-numeric columns. skip_blank_lines : bool, default True If True, skip over blank lines rather than interpreting as NaN values. parse_dates : bool or list of int or names or list of lists or dict, default False The behavior is as follows: * boolean. If True -> try parsing the index. * list of int or names. e.g. If [1, 2, 3] -> try parsing columns 1, 2, 3 each as a separate date column. * list of lists. e.g. If [[1, 3]] -> combine columns 1 and 3 and parse as a single date column. * dict, e.g. {'foo' : [1, 3]} -> parse columns 1, 3 as date and call result 'foo' If a column or index cannot be represented as an array of datetimes, say because of an unparseable value or a mixture of timezones, the column or index will be returned unaltered as an object data type. For non-standard datetime parsing, use ``pd.to_datetime`` after ``pd.read_csv``. To parse an index or column with a mixture of timezones, specify ``date_parser`` to be a partially-applied :func:`pandas.to_datetime` with ``utc=True``. See :ref:`io.csv.mixed_timezones` for more. Note: A fast-path exists for iso8601-formatted dates. infer_datetime_format : bool, default False If True and `parse_dates` is enabled, pandas will attempt to infer the format of the datetime strings in the columns, and if it can be inferred, switch to a faster method of parsing them. In some cases this can increase the parsing speed by 5-10x. keep_date_col : bool, default False If True and `parse_dates` specifies combining multiple columns then keep the original columns. date_parser : function, optional Function to use for converting a sequence of string columns to an array of datetime instances. The default uses ``dateutil.parser.parser`` to do the conversion. Pandas will try to call `date_parser` in three different ways, advancing to the next if an exception occurs: 1) Pass one or more arrays (as defined by `parse_dates`) as arguments; 2) concatenate (row-wise) the string values from the columns defined by `parse_dates` into a single array and pass that; and 3) call `date_parser` once for each row using one or more strings (corresponding to the columns defined by `parse_dates`) as arguments. dayfirst : bool, default False DD/MM format dates, international and European format. cache_dates : bool, default True If True, use a cache of unique, converted dates to apply the datetime conversion. May produce significant speed-up when parsing duplicate date strings, especially ones with timezone offsets. .. versionadded:: 0.25.0 iterator : bool, default False Return TextFileReader object for iteration or getting chunks with ``get_chunk()``. chunksize : int, optional Return TextFileReader object for iteration. See the `IO Tools docs <https://pandas.pydata.org/pandas-docs/stable/io.html#io-chunking>`_ for more information on ``iterator`` and ``chunksize``. compression : {'infer', 'gzip', 'bz2', 'zip', 'xz', None}, default 'infer' For on-the-fly decompression of on-disk data. If 'infer' and `filepath_or_buffer` is path-like, then detect compression from the following extensions: '.gz', '.bz2', '.zip', or '.xz' (otherwise no decompression). If using 'zip', the ZIP file must contain only one data file to be read in. Set to None for no decompression. thousands : str, optional Thousands separator. decimal : str, default '.' Character to recognize as decimal point (e.g. use ',' for European data). lineterminator : str (length 1), optional Character to break file into lines. Only valid with C parser. quotechar : str (length 1), optional The character used to denote the start and end of a quoted item. Quoted items can include the delimiter and it will be ignored. quoting : int or csv.QUOTE_* instance, default 0 Control field quoting behavior per ``csv.QUOTE_*`` constants. Use one of QUOTE_MINIMAL (0), QUOTE_ALL (1), QUOTE_NONNUMERIC (2) or QUOTE_NONE (3). doublequote : bool, default ``True`` When quotechar is specified and quoting is not ``QUOTE_NONE``, indicate whether or not to interpret two consecutive quotechar elements INSIDE a field as a single ``quotechar`` element. escapechar : str (length 1), optional One-character string used to escape other characters. comment : str, optional Indicates remainder of line should not be parsed. If found at the beginning of a line, the line will be ignored altogether. This parameter must be a single character. Like empty lines (as long as ``skip_blank_lines=True``), fully commented lines are ignored by the parameter `header` but not by `skiprows`. For example, if ``comment='#'``, parsing ``#empty\na,b,c\n1,2,3`` with ``header=0`` will result in 'a,b,c' being treated as the header. encoding : str, optional Encoding to use for UTF when reading/writing (ex. 'utf-8'). `List of Python standard encodings <https://docs.python.org/3/library/codecs.html#standard-encodings>`_ . dialect : str or csv.Dialect, optional If provided, this parameter will override values (default or not) for the following parameters: `delimiter`, `doublequote`, `escapechar`, `skipinitialspace`, `quotechar`, and `quoting`. If it is necessary to override values, a ParserWarning will be issued. See csv.Dialect documentation for more details. error_bad_lines : bool, default True Lines with too many fields (e.g. a csv line with too many commas) will by default cause an exception to be raised, and no DataFrame will be returned. If False, then these "bad lines" will dropped from the DataFrame that is returned. warn_bad_lines : bool, default True If error_bad_lines is False, and warn_bad_lines is True, a warning for each "bad line" will be output. delim_whitespace : bool, default False Specifies whether or not whitespace (e.g. ``' '`` or ``' '``) will be used as the sep. Equivalent to setting ``sep='\s+'``. If this option is set to True, nothing should be passed in for the ``delimiter`` parameter. low_memory : bool, default True Internally process the file in chunks, resulting in lower memory use while parsing, but possibly mixed type inference. To ensure no mixed types either set False, or specify the type with the `dtype` parameter. Note that the entire file is read into a single DataFrame regardless, use the `chunksize` or `iterator` parameter to return the data in chunks. (Only valid with C parser). memory_map : bool, default False If a filepath is provided for `filepath_or_buffer`, map the file object directly onto memory and access the data directly from there. Using this option can improve performance because there is no longer any I/O overhead. float_precision : str, optional Specifies which converter the C engine should use for floating-point values. The options are `None` for the ordinary converter, `high` for the high-precision converter, and `round_trip` for the round-trip converter. Returns ------- DataFrame or TextParser A comma-separated values (csv) file is returned as two-dimensional data structure with labeled axes. See Also -------- DataFrame.to_csv : Write DataFrame to a comma-separated values (csv) file. read_csv : Read a comma-separated values (csv) file into DataFrame. read_fwf : Read a table of fixed-width formatted lines into DataFrame. Examples -------- >>> pd.read_csv('data.csv') # doctest: +SKIP File: ~/opt/anaconda3/lib/python3.8/site-packages/pandas/io/parsers.py Type: function
Boolean masking¶
Boolean masking is a very powerful technique that is the key to fast and efficient querying of datasets.
A boolean mask has the same size of our data, but its values can be either True or False. The mask is "overlaid" on the data to select ONLY the values which correspond to True. Here is a visual explanation by Matt Eding on a 2D dataset:
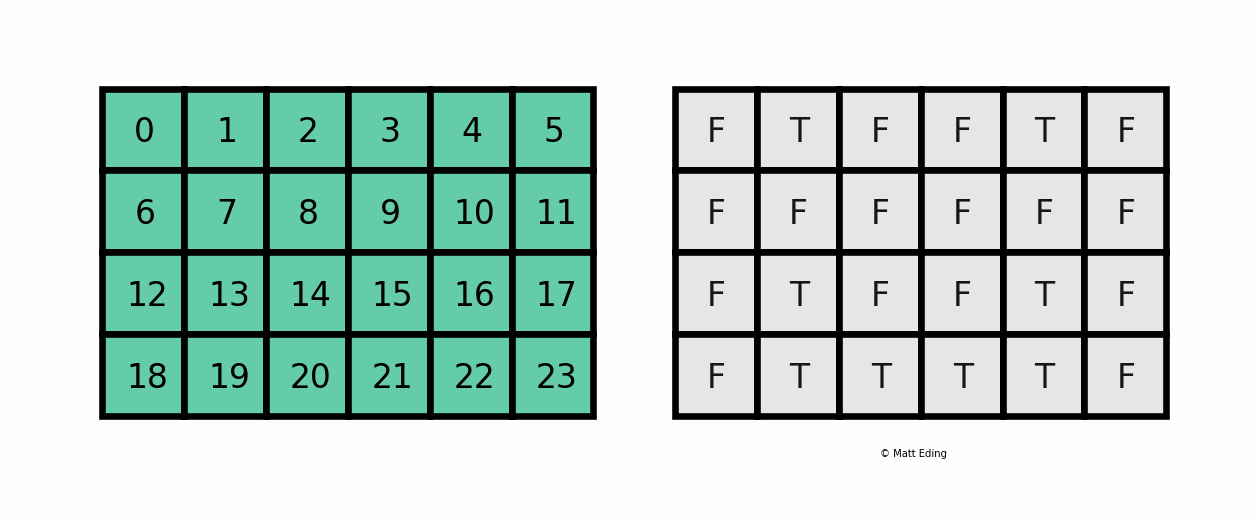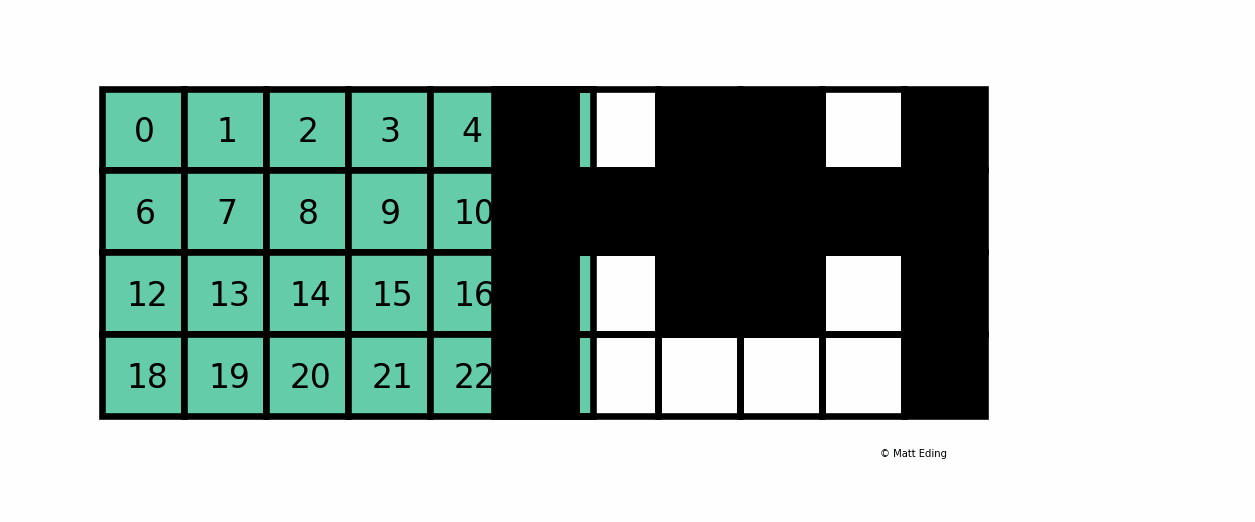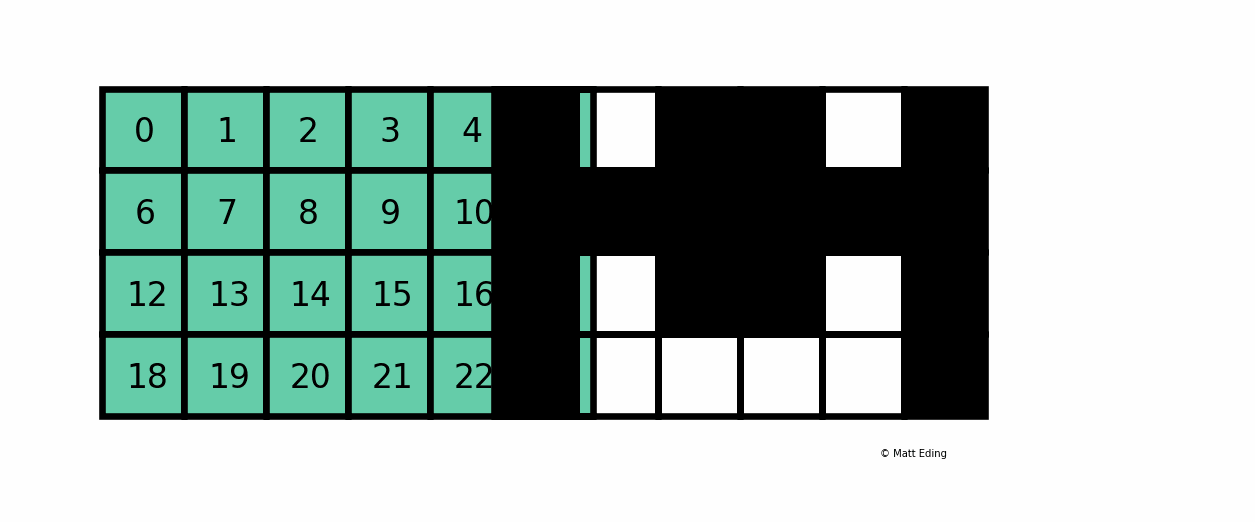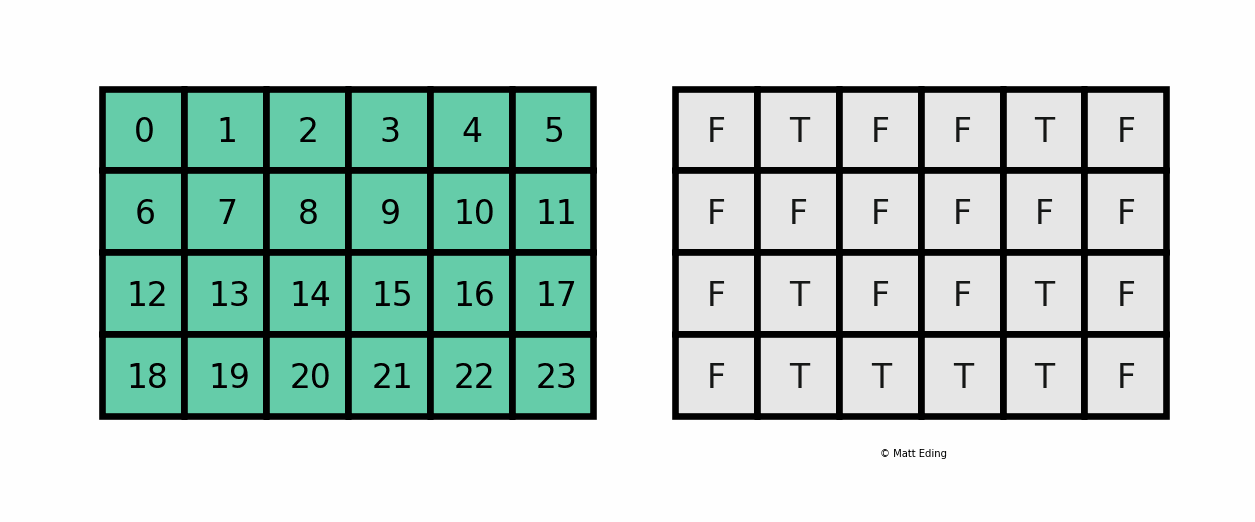
Masking Series¶
Boolean masking can be done in two steps:
- Generate the mask (using vectorised operations);
- Apply the mask to the data.
# As an example :
y = pd.Series([100, 101, 102, 10])
- Generate the mask
mask = y > 10
mask
0 True 1 True 2 True 3 False dtype: bool
NOTE that the boolean mask:
- contains
Truefor all values of theSeriesthat match the criterion, andFalseotherwise; - is of the same size as the
Series.
- Apply the mask
We apply (overlay) this mask on the data by using .loc[].
y.loc[mask]
0 100 1 101 2 102 dtype: int64
Exercise 8¶
Remember our x Series from before? Extract from x the elements as specified below:
- All elements greater than 4;
- All elements smaller than 4.
# Solve here
mask = x > 4
x.loc[mask]
mask = x < 4
x.loc[mask]
a 0 b 1 c 2 d 3 dtype: int64
Masking DataFrames¶
It is possible to create 2D boolean masks and apply them to DataFrames (as in the visual exemple above); but most often, we want to filter rows of a DataFrame using a mask generated based on values of a specific column. Here is a visual explanation:
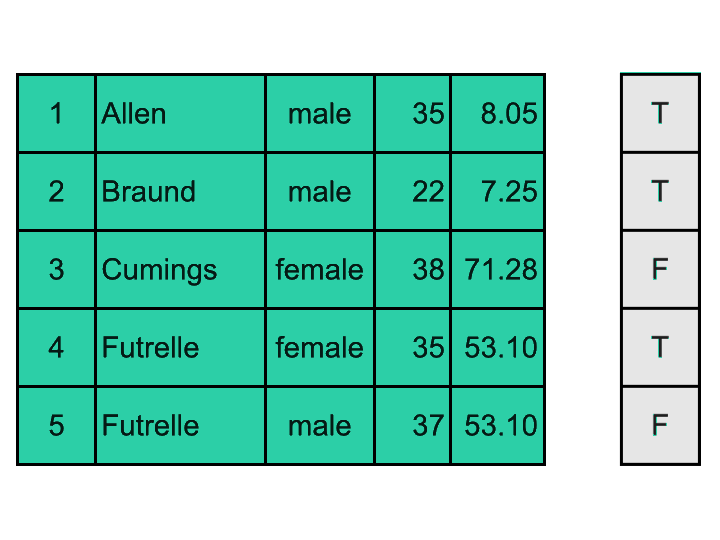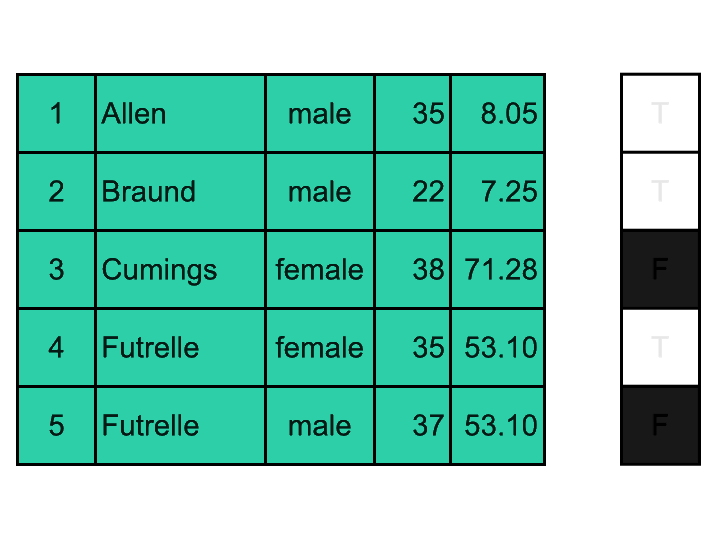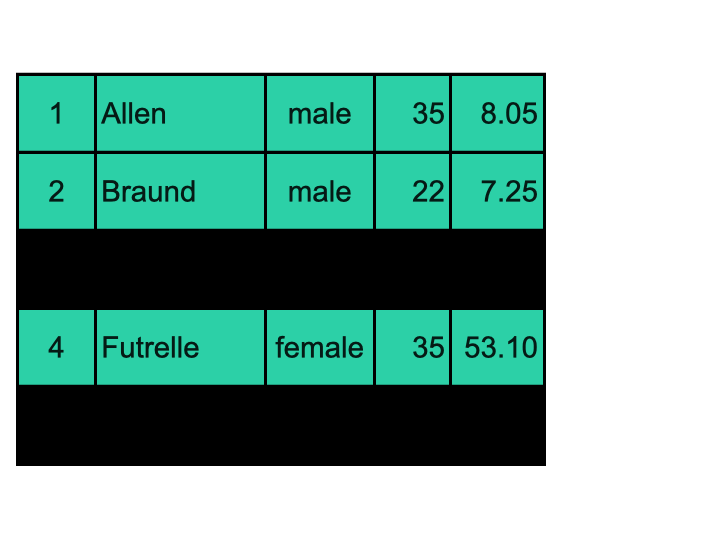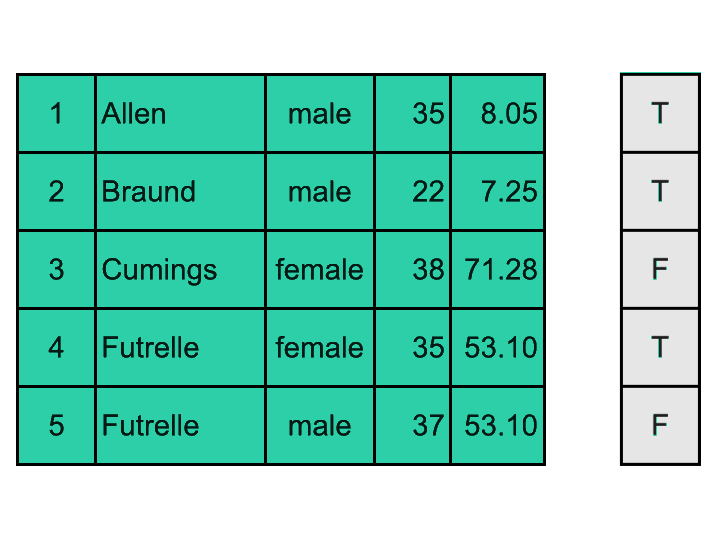
We are going to:
- Generate a mask using vectorised operations on a column;
- Apply the mask using
.loc[].
- Generate the mask
mask = titanic.loc[:, "age"] > 30
NOTE that the boolean mask:
- contains
Truefor all values of the column that match the criterion, andFalseotherwise; - is of the same size as a column of the
DataFrame.
- Apply the mask
We apply (overlay) this mask on the data by using .loc[]. The result is filtering only a subset of rows of the DataFrame (for example, only passengers whose age is greater than 30).
titanic.loc[mask, :]
| sex | age | fare | survived | |
|---|---|---|---|---|
| name | ||||
| Allen | male | 35 | 8.05 | 0 |
| Cumings | female | 38 | 71.28 | 1 |
| Futrelle | female | 35 | 53.10 | 1 |
| Futrelle | male | 37 | 53.10 | 0 |
Combining masks¶
A very powerful technique is to perform complex queryies by combine multiple boolean masks using the logical operators & (and), | (or), ~ (not). As you might expect, boolean masks need to be the same size to be combined.
- Generate the mask
age_mask = titanic.loc[:, "age"] > 35
fare_mask = titanic.loc[:, "fare"] > 10
mask = age_mask & fare_mask
- Apply the mask
titanic.loc[mask, :]
| sex | age | fare | survived | |
|---|---|---|---|---|
| name | ||||
| Cumings | female | 38 | 71.28 | 1 |
| Futrelle | male | 37 | 53.10 | 0 |
Exercise 9¶
Extract from the titanic Dataframe the elements as specified below:
- Passengers that payed less than 10;
- Passengers that are 35 or older;
- Just the sex of passengers that survived;
- All male passengers;
- Passengers that survived and are younger than 30;
- Passengers that payed more than 10 and whose age is not even;
# 1.
fare_mask = titanic.loc[:,"fare"] < 10
titanic.loc[fare_mask, :]
# 2.
age_mask = titanic.loc[:,"age"] >= 35
titanic.loc[age_mask, :]
# 3.
survive_mask = titanic.loc[:,"survived"] == 1
titanic.loc[survive_mask, "sex"]
# 4.
sex_mask = titanic.loc[:,"sex"] == "male"
titanic.loc[sex_mask, :]
# 5.
age_mask = titanic.loc[:,"age"] < 30
survive_mask = titanic.loc[:,"survived"] == 1
mask = age_mask & survive_mask
titanic.loc[mask, :]
# 6.
fare_mask = titanic.loc[:,"fare"] > 10
age_mask = titanic.loc[:,"age"] % 2 != 0
mask = age_mask & fare_mask
titanic.loc[mask, :]
| sex | age | fare | survived | |
|---|---|---|---|---|
| name | ||||
| Futrelle | female | 35 | 53.1 | 1 |
| Futrelle | male | 37 | 53.1 | 0 |
Extra Exercises and mini-Projects¶
Exercise 10: Querying¶
You have the following Series:
>>> series = pd.Series(range(5, 20))
Extract values from this Series as follows:
- The first value;
- The last value;
- The last 5 values;
- Every fifth value starting at the beginning;
- Values greater than 6;
- Values smaller than 5;
- Values greater than 10 that are multiples of 3.
series = pd.Series(range(5, 20))
series.iloc[0]
series.iloc[-1]
series.iloc[::5]
series.loc[series > 6]
series.loc[series < 5]
series.loc[(series > 10) & (series%3 == 0)] # Note the parentheses (...)
Exercise 11: Multiples of 5¶
Write function only_multiples_of_five(s) that returns those elements of Series s that are multiples of 5.
For example:
>>> series = pd.Series([1, 9, 8, 6, 10, 5, 1, 2, 10, 1])
>>> only_multiples_of_five(series)
4 10
5 5
8 10
dtype: int64
def only_multiples_of_five(s):
return s.loc[s%5==0]
series = pd.Series([1, 9, 8, 6, 10, 5, 1, 2, 10, 1])
only_multiples_of_five(series)
Exercise 12: Euclidean distance¶
Write a function euclidean_distance(s1, s2) to compute the Euclidean distance (https://en.wikipedia.org/wiki/Euclidean_distance) between two given Series.
>>> series1 = pd.Series(range(1, 11))
>>> series2 = pd.Series([11, 8, 7, 5, 6, 5, 3, 4, 7, 1])
>>> euclidean_distance(series1, series2)
16.492422502470642
Exercise 13: More titanic¶
Use the titanic DataFrame to calculate the mean price payed by people who survived, and by people who died.
titanic = pd.read_csv("https://marcopasi.github.io/physenbio_pyDAV/data/titanic_tiny.csv", index_col=0)
survived = titanic.loc[:, "survived"] == 1
print(f"Survivors paid on average {titanic.loc[survived, 'fare'].mean()}")
print(f"Non-survivors paid on average {titanic.loc[~survived, 'fare'].mean()}") # Note ~ = not
Exercise 14: Exams¶
- Based on the variables defined below, create a
DataFramenamedexamthat resembles the following table, that contains the best score for an exam (over multiple attempts) for some students.
- Based on the variables defined below, create a
| index | name | score | attempts |
|---|---|---|---|
| a | Anastasia | 12.5 | 1 |
| b | Dima | 9.0 | 3 |
| c | Katherine | 16.5 | 2 |
| d | James | -1.0 | 3 |
| e | Emily | 9.0 | 2 |
| f | Michael | 20.0 | 3 |
| g | Matthew | 14.5 | 1 |
| h | Laura | -1.0 | 1 |
| i | Kevin | 8.0 | 2 |
| j | Jonas | 19.0 | 1 |
names = ['Anastasia', 'Dima', 'Katherine', 'James', 'Emily', 'Michael', 'Matthew', 'Laura', 'Kevin', 'Jonas']
scores = [12.5, 9, 16.5, -1, 9, 20, 14.5, -1, 8, 19]
attempts = [1, 3, 2, 3, 2, 3, 1, 1, 2, 1]
exam = pd.DataFrame({"name": names, "score": scores, "attempts": attempts},
index=list("abcdefghij"))
exam
From the exam DataFrame extract the following:
- The first 3 rows;
- The
nameandscoreof all students;
- The
- The names of all columns, and the labels of all rows;
exam.iloc[:3,:]
exam.loc[:, ["name", "score"]]
exam.columns
exam.index
Modify the exam DataFrame as indicated in the following:
- Change the score of Katherine to 17;
- Change the number of attempts of James to 1;
exam.loc["c", "score"] = 17
exam.loc["d", "attempts"] = 1
exam
- Select students who have attempted the exam more than once.
exam.loc[exam.loc[:, "attempts"] > 1, :]
Use the exam DataFrame to calculate the following:
- The total number of attempts performed by all students;
- The mean score of all students;
exam.loc[:, "attempts"].sum()
exam.loc[:, "score"].mean()
- The passing threshold for this exam is 10. Add a column
qualifythat isTrueif the student passes the exam, andFalseotherwise. Consider that ascoreof -1 indicates the student was absent (impossible to assign ascore).
- The passing threshold for this exam is 10. Add a column
exam.loc[:, "qualify"] = exam.loc[:, "score"] > 10
exam
Project 1: Flower shop¶
You are hired as data analyst for a chain of gardening shops. They provide you with their inventory in the form of a CSV file:
https://marcopasi.github.io/physenbio_pyDAV/data/inventory.csv
The questions below will guide you through the analysis of this dataset.
- Import the dataset from the
csvfile
- Inspect the first 10 rows of the inventory
- The first 10 rows represent data from your Staten Island location. Find what products are sold at your Staten Island location (extract the
product_descriptionfor these products)
- Find what types of seeds are sold at the Brooklyn location: select all rows where
locationis equal toBrooklynandproduct_typeis equal toseeds
- Add a column to the inventory called
in_stockwhich isTrueifquantityis greater than 0 andFalseifquantityequals 0.
- Calculate the total value of the current inventory.
HINT: first create a new column called total_value that is equal to price multiplied by quantity.
Project 2: Euro 2012¶

You are writing an article with an in-depth analysis of the 2012 edition of the UEFA European Championship. You are provided detailed data about the championship in the form of a CSV file:
https://marcopasi.github.io/physenbio_pyDAV/data/Euro_2012_stats_TEAM.csv
The questions below will guide you through the data analysis.
- Import the dataset from the
csvfile
- Inspect the first 10 lines of the data
- Select only the Goal column
- How many team participated in the Euro2012?
- What is the number of columns in the dataset?
- Extract only the columns
Team,Yellow CardsandRed Cardsand assign them to a new dataframe
- Calculate the mean number of Yellow Cards given per Team
- Calculate the total number of cards (Yellow and Red) given per Team
- Filter teams that scored more than 6 goals
- Select the first 7 columns
- Select all columns except the last 3
License¶

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
